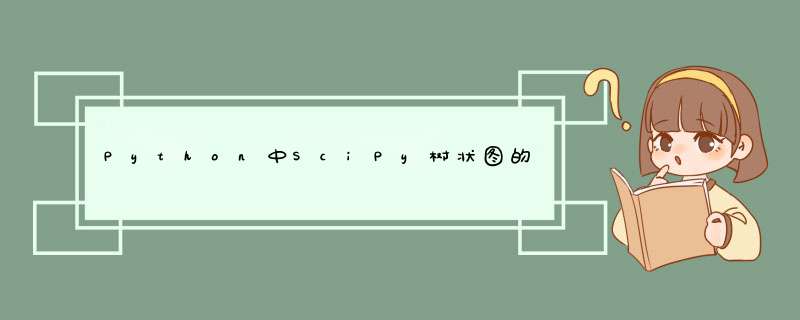
这是一个使用返回矩阵
Z的解决方案
linkage()(早有描述,但在文档中有所隐藏)和
link_color_func:
# see question for pre prior to "color mapping"# Color mappingdflt_col = "#808080" # Unclustered grayD_leaf_colors = {"attr_1": dflt_col, "attr_4": "#B061FF", # Cluster 1 indigo "attr_5": "#B061FF", "attr_2": "#B061FF", "attr_8": "#B061FF", "attr_6": "#B061FF", "attr_7": "#B061FF", "attr_0": "#61ffff", # Cluster 2 cyan "attr_3": "#61ffff", "attr_9": "#61ffff", }# notes:# * rows in Z correspond to "inverted U" links that connect clusters# * rows are ordered by increasing distance# * if the colors of the connected clusters match, use that color for linklink_cols = {}for i, i12 in enumerate(Z[:,:2].astype(int)): c1, c2 = (link_cols[x] if x > len(Z) else D_leaf_colors["attr_%d"%x] for x in i12) link_cols[i+1+len(Z)] = c1 if c1 == c2 else dflt_col# DendrogramD = dendrogram(Z=Z, labels=DF_dism.index, color_threshold=None, leaf_font_size=12, leaf_rotation=45, link_color_func=lambda x: link_cols[x])这里的输出:
欢迎分享,转载请注明来源:内存溢出

 微信扫一扫
微信扫一扫
 支付宝扫一扫
支付宝扫一扫
评论列表(0条)