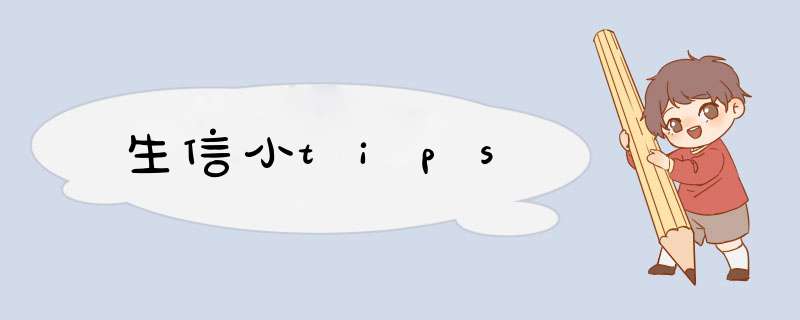
fasta2phy:
cat 4.4Dsites.pl.connect4Dsites.fa | tr '\n' '\t' | \
pipe pipe>sed 's/>/\n/g' |
pipe pipe pipe>sed 's/\t//' | sed 's/\t//g' |
pipe pipe pipe pipe pipe>awk 'NF >0' >4.4Dsites.pl.connect4Dsites.fa.tmp
awk '{print " "NR" "length($2)}' 4.4Dsites.pl.connect4Dsites.fa.tmp |
pipe>tail -n 1 | \
pipe pipe>cat - 4.4Dsites.pl.connect4Dsites.fa.tmp >4.4Dsites.pl.connect4Dsites.phy
fasteprr 分型报错!
zcat input.vcf.gz | perl -pe "s/\s.:/\t./.:/g" | bgzip -c >output.vcf.gz
Structure
For population structure
analyses, we discarded SNPs with missing rate >20%
and minor allele frequency (MAF) <5%. We also excluded
highly correlated SNPs by performing an LD-based SNP
pruning process in PLINK v1.90
Mental test
VEGAN(R package)
质控软件
trimmomatic (需要知道接头序列)
FSC
We excluded SNPs from three genomic regions
under long-term balancing selection (see the “Balancing
selection in B. stricta genomes” section) and only used
fourfold degenerate sites and intergenic regions, because
they are less affected by selection.
To account for the influence of effective population
size on estimated ρ, we divided ρ by diversity (π) in
each 20-kb window following Wang et al. [4] and compared
ρ/π between islands and the rest of the genome.
提取奇数行 sed -n '1~2p' a.txt
提取偶数行 sed -n '0~2p' a.txt
shuf -n100 filename
sort -R filename | head -n100
awk '{x+=1}{print 3"\t"$4}' test.map >map
其中 : tree 文件夹中: estimated_gene_trees.tree 基因树
estimated_species_tree.tree 并联树
name 文件:
singularity sif 转 sandbox
几款构建祖先染色体的软件
一到一还谈什么随机?
如果是一个文件夹中的文件随机复制到N个文件夹中的一个,那么可以把N个文件夹的路径放到一个数组变量中,并用 $RANDOM 来产生随机数,对数组长度取余后作为下标。
#!/bin/bashdests=( /dest/a /dest/b /dest/c /destd)
for f in * do
cp "$f" ${dests[((RANDOM%${#dests[@]}))]
done
#!/bin/bashsort -R a.txt | head -20000
##
sort随机排序,然后取前20000,实现出来就是随机抽取20000.
欢迎分享,转载请注明来源:内存溢出

 微信扫一扫
微信扫一扫
 支付宝扫一扫
支付宝扫一扫
评论列表(0条)