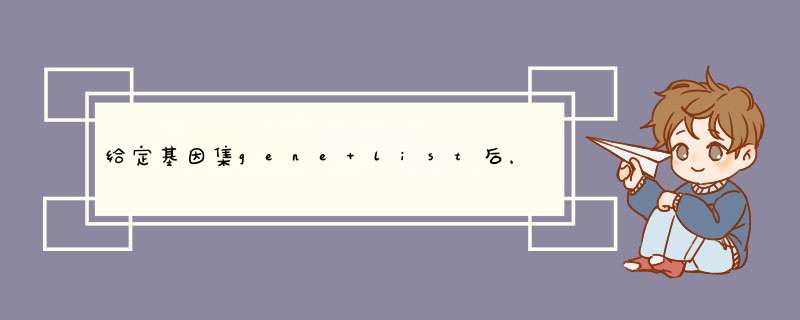
一.把准备好的gene名字复制到txt文件中(此时是symbol格式的gene名,如下,得到gene.txt
二.安装所需包
if (!requireNamespace("BiocManager", quietly = TRUE))
install.packages("BiocManager")
BiocManager::install("clusterProfiler")
library(clusterProfiler)
BiocManager::install("AnnotationHub")
library(AnnotationHub)
BiocManager::install("org.Hs.eg.db")
library(org.Hs.eg.db)
library(ggplot2)
BiocManager::install("DOSE")
library(DOSE)三.ID转换
#1 读取第一步中的txt文件
go_ythdf2 <- read.table("gene.txt",sep=" ")#读取
go_ythdf2 <- t(go_ythdf2)#从列 变为行
#2 读取db包
keytypes(org.Hs.eg.db)
#3 将gene_symbol转换为ENSEMBL
go_ythdf2_id_trance <- bitr(go_ythdf2,fromType = "SYMBOL",toType = "ENSEMBL",OrgDb = "org.Hs.eg.db",drop = T)
#4 提取ENSEMBL的信息
f <- as.data.frame(go_ythdf2_id_trance[,2])
colnames(f)[1] <- ''
colnames(f) <- "V1"
EG2Ensembl=toTable(org.Hs.egENSEMBL)
f=f$V1
geneLists=data.frame(ensembl_id=f)
results=merge(geneLists,EG2Ensembl,by='ensembl_id',all.x=T)
id=na.omit(results$gene_id)四.GO分析
#1 GO分析
All <- enrichGO(OrgDb="org.Hs.eg.db", gene = id, ont = "ALL", readable= TRUE)
save(All,file = 'All.rda')
#如果要单独绘制BP,CC,MF,把ont="ALL"修改成自己想要的类型即可
#2 画图,气泡图
#2.1 气泡图,显示前10项,标题为“Enrichment GO Up-Gene”
dotplot(All,showCategory=10,title="Enrichment GO Up-Gene")+
scale_color_gradient(low = "red", high = "blue")+
theme_bw()
#2.2 柱状图,显示前20项,标题为“Enrichment GO Top20”
barplot(All, showCategory=20,title="EnrichmentGO")
# 2.3 BP,MF,CC分别显示(要加载DOSE包)
p1 <- dotplot(All,split="ONTOLOGY",title="Enrichment GO Up-Gene")+ facet_grid(ONTOLOGY~.,scale="free")+
theme_bw()
p1
ggsave('GO.pdf',p1,width = 8,height = 10)
# 只显示8个通路
# p3 <- dotplot(All,split="ONTOLOGY",showCategory=8,title="Enrichment GO Up-Gene")+ facet_grid(ONTOLOGY~.,scale="free")+
# theme_bw()
# p3
# ggsave('GO8.pdf',p3,width = 8,height = 10)
#三个单独出图
BP <- enrichGO(OrgDb="org.Hs.eg.db", gene = id, ont = "BP", readable= TRUE)
p10 <- dotplot(BP,showCategory=10,title="Enrichment GO Up-Gene-BP")+
scale_color_gradient(low = "red", high = "blue")+
theme_bw()
ggsave('BF10.pdf',p10,width = 8,height = 6)
MF <- enrichGO(OrgDb="org.Hs.eg.db", gene = id, ont = "MF", readable= TRUE)
p12 <- dotplot(MF,showCategory=10,title="Enrichment GO Up-Gene-MF")+
scale_color_gradient(low = "red", high = "blue")+
theme_bw()
ggsave('MF10.pdf',p12,width = 8,height = 6)
CC <- enrichGO(OrgDb="org.Hs.eg.db", gene = id, ont = "CC", readable= TRUE)
p14 <- dotplot(CC,showCategory=10,title="Enrichment GO Up-Gene-CC")+
scale_color_gradient(low = "red", high = "blue")+
theme_bw()
ggsave('CC10.pdf',p14,width = 8,height = 6)
五、KEGG分析
#1KEGG分析
KEGG <- enrichKEGG(gene= id, organism = 'hsa', pvalueCutoff = 0.05)
#2 画图,第一种做法,气泡图,设置字体大小
dotplot(KEGG,font.size=12)
#3 画图,第二种做法,柱状图
barplot(KEGG,font.size=8)
#4 画图,第三种做法,泡泡图
dotplot(KEGG,showCategory=10,title="Enrichment KEGG Top10")
#4.1 调节标题,字体,泡泡大小
p2 <- dotplot(KEGG, font.size=8, showCategory=10, title="Enrichment KEGG Top10") + scale_size(rang=c(5.20))
p2
ggsave('KEGG10.pdf',p2,width = 6,height = 6)
这样分析就结束啦,放上gene list练手
链接:https://pan.baidu.com/s/1wlwV6ItpLx_oIJzypI2crQ
提取码:mr4o
欢迎分享,转载请注明来源:内存溢出

 微信扫一扫
微信扫一扫
 支付宝扫一扫
支付宝扫一扫
评论列表(0条)